In step 1 of IBD segment extraction from FABIA models as
described in subsection
``Extraction of IBD Segments from FABIA Models'' of the main
manuscript,
the likelihood of observing identity by state (IBS)
without IBD and, therefore, without recombination, is computed.
If ![]() is the minor allele frequency (MAF) for one SNV,
the probability of observing the minor allele of this SNV in
all
is the minor allele frequency (MAF) for one SNV,
the probability of observing the minor allele of this SNV in
all ![]() individuals is
individuals is ![]() .
We assumed that all SNVs have the same MAF
.
We assumed that all SNVs have the same MAF ![]() -- in the
experiments we used the average MAF.
According to the main manuscript,
the probability of observing
-- in the
experiments we used the average MAF.
According to the main manuscript,
the probability of observing ![]() or more counts of model
SNVs by chance in an interval of
or more counts of model
SNVs by chance in an interval of ![]() SNVs is
SNVs is
If we try to minimize this probability, we observe a trade-off
between small average minor allele frequency (MAF) ![]() and the number
and the number ![]() of individuals that share an IBD segment.
For a small IBS likelihood, the average MAF
of individuals that share an IBD segment.
For a small IBS likelihood, the average MAF
![]() should be small and
should be small and ![]() large.
However, increasing
large.
However, increasing ![]() also increases
the lower bound
also increases
the lower bound
![]() on
on ![]() .
If we assume minimal
.
If we assume minimal
![]() , then
above probability Eq. (2) is governed by the term
, then
above probability Eq. (2) is governed by the term
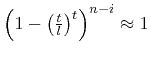 .
The function
.
The function
To avoid IBS without IBD by recombination,
we focused on rare SNVs (see main manuscript).
IBD can be distinguished from IBS without
IBD by rare alleles, because for them two independent origins are
unlikely, so IBS generally implies IBD, which is not true for
common alleles (3, chapter 15.3, p. 441).
Hence, the minimum ![]() will not be attained with rare SNVs.
For fewer individuals
will not be attained with rare SNVs.
For fewer individuals ![]() than the minimum
than the minimum ![]() ,
the function
,
the function
![]() is decreasing.
This justifies that more individuals give more evidence for IBD
because random minor allele sharing (IBS without IBD) is less likely.
is decreasing.
This justifies that more individuals give more evidence for IBD
because random minor allele sharing (IBS without IBD) is less likely.